Biomedical data fusion via sequence alignment—Part 2
This is the second part of my discussion on comparing and merging multiple time-series data sequences together. In Part 1, I briefly discussed how we can use Dynamic Programming to align two data sequences. In this post, I will describe a fast algorithm to extend sequence alignment to more than two sequences.
It’s easy to show that the time complexity of the Needleman-Wunsch algorithm applied to two sequences with approximately same length is . The algorithm can be extended directly to simultaneously align sequences, but this comes with a huge cost: an exponential time complexity of .
So what can we do? This is another instance where use of an appropriate data structure can help algorithm design to make impracticals feasible. Let me take a quick detour and briefly describe the capabilities of this data-structure first. If you are not familiar with data structures and their purposes in general, you should familiarize yourself with this concept.
Union-find is a data structure that keeps track of partitioning of a set S with size into a disjoint set of groups with fast implementation of the following operations:
find(x): returns the name of the group that contains element in ,union(G1,G2): merges two groups G1 and G2 in , andmakeUnionFind(S): initializes the data structure with each element of set S in a separate group in .
Back to the problem at hand, now that we know the properties of union-find, we use pairwise sequence alignment (between every distinct pair of sequences), and use a union-find data structure to keep track of the alignments. I’ll discuss later in this post why using this technique will hugely decrease the time complexity of multiple sequence alignment.
So here is what I’m going to do: I’ll first describe the algorithm by writing the pseudocode, and then provide its Python implementation for those who would like to try it out for themselves. We need a little bit of extra notation to describe the pseudocode. Let denote the set of data sequences, each containing elements (elements being time points, in case of time-series data). Also let refer to the th element of th sequence.
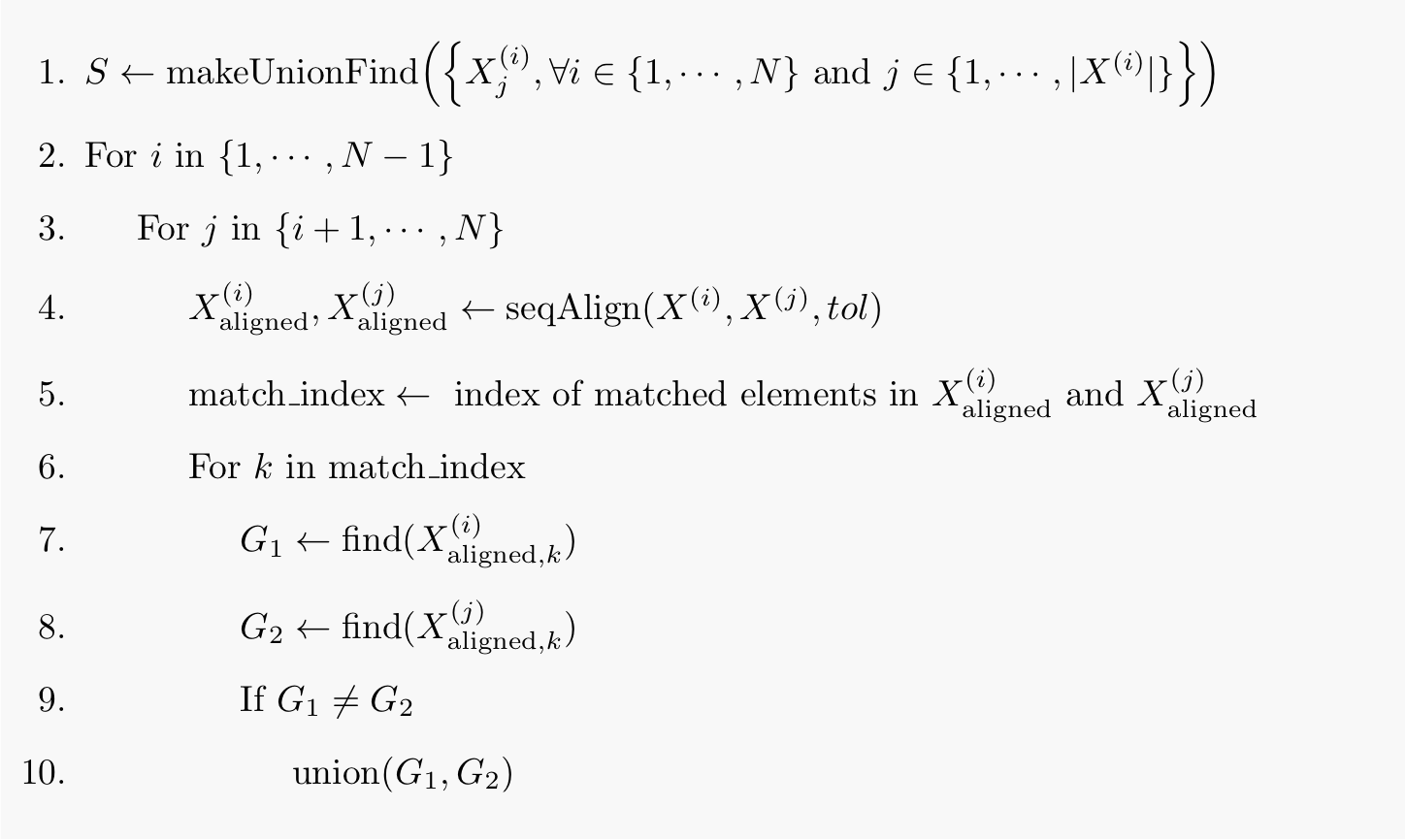
At first, the union-find data structure is initialized with each element of each sequence in its own separate group. Next, every distinct pair of data sequences are aligned together, and their matched elements are put in the same group. At the end of pairwise sequence alignments, the union-find data structure contains the alignment information. Using the time complexities of pairwise sequence alignment and different union-find operations, it is easy to show that the time complexity of multiple sequence alignment using a union-find data structure is . This is a huge gain over the exponential time complexity described at the beginning of this post.
With the above description, it should be fairly easy to go through the Python implementation and find correspondences with the pseudocode. I have used this Python implementation of a union-find data structure.
from Classes import Node, UnionFind
def seqVoting(seq_list):
# Creating the node list
node_list = UnionFind()
for i in range(len(seq_list)):
for item in seq_list[i]:
n = Node((i,item))
node_list.addnode(n)
for i in range(len(seq_list)-1):
for j in range(i+1,len(seq_list)):
seq1_aligned,seq2_aligned = seqAlign(seq_list[i],
seq_list[j], tol)
matchUnion(seq1_aligned,seq2_aligned,i,j,node_list)
return node_listseqAlign is the code for pairwise sequence alignment, described in the previous post, and matchUnion which transfers the alignment information to the union-find data structure has the following implementation:
import numpy as np
def matchUnion(seq1_aligned, seq2_aligned, id1, id2, node_list):
nan_ind = np.r_[np.where(np.isnan(seq1_aligned))[0],
np.where(np.isnan(seq2_aligned))[0]]
non_nan_ind = [i for i in range(len(seq1_aligned))
if i not in nan_ind]
for i in non_nan_ind:
n1 = node_list.getNode((id1, seq1_aligned[i]))
n2 = node_list.getNode((id2, seq2_aligned[i]))
if node_list.findset(n1) != node_list.findset(n2):
node_list.union(n2, n1)In summary, the algorithms described in the last two posts provide an efficient way to merge information of multiple time-series data. It should be evident by now that aligning two data sequences is an important first step for any data merging/fusion algorithm. Once data sequences are aligned, different deterministic or probabilistic merging algorithms (such as majority voting, mean or median data fusion, or Bayesian voting) can be applied to the aligned sequences to further analyze the data.
There are numerous applications for this technique, many of which I am not aware of, but you can find one application for this algorithm in this paper, which shows the importance of sequence alignment for comparing ECG delineation results with their corresponding gold standards.